Parameterizations¶
In this notebook, we’ll review tools for defining, running, and comparing subgrid parameterizations.
[1]:
import numpy as np
import pandas as pd
pd.set_option('display.max_columns', 10)
import pyqg
import pyqg.diagnostic_tools
import matplotlib.pyplot as plt
%matplotlib inline
Run baseline high- and low-resolution models¶
To illustrate the effect of parameterizations, we’ll run two baseline models:
a low-resolution model without parameterizations at
nx=64resolution (where \(\Delta x\) is larger than the deformation radius \(r_d\), preventing the model from fully resolving eddies),a high-resolution model at
nx=256resolution (where \(\Delta x\) is ~4x finer than the deformation radius, so eddies can be almost fully resolved).
[2]:
%%time
year = 24*60*60*360.
base_kwargs = dict(dt=3600., tmax=5*year, tavestart=2.5*year, twrite=25000)
low_res = pyqg.QGModel(nx=64, **base_kwargs)
low_res.run()
INFO: Logger initialized
INFO: Step: 25000, Time: 9.00e+07, KE: 4.14e-04, CFL: 0.042
CPU times: user 14.3 s, sys: 23.6 s, total: 38 s
Wall time: 10.8 s
[3]:
%%time
high_res = pyqg.QGModel(nx=256, **base_kwargs)
high_res.run()
INFO: Logger initialized
INFO: Step: 25000, Time: 9.00e+07, KE: 4.62e-04, CFL: 0.217
CPU times: user 3min 47s, sys: 5min 43s, total: 9min 30s
Wall time: 2min 55s
Run Smagorinsky and backscatter parameterizations¶
Now we’ll run two types of parameterization: one from Smagorinsky 1963 which models an effective eddy viscosity from subgrid stress, and one adapted from Jansen and Held 2014 and Jansen et al. 2015, which reinjects a fraction of the energy dissipated by Smagorinsky back into larger scales:
[4]:
def run_parameterized_model(p):
model = pyqg.QGModel(nx=64, parameterization=p, **base_kwargs)
model.run()
return model
[5]:
%%time
smagorinsky = run_parameterized_model(
pyqg.parameterizations.Smagorinsky(constant=0.08))
INFO: Logger initialized
INFO: Step: 25000, Time: 9.00e+07, KE: 3.37e-04, CFL: 0.043
CPU times: user 39.3 s, sys: 1min, total: 1min 39s
Wall time: 30.3 s
[8]:
%%time
backscatter = run_parameterized_model(
pyqg.parameterizations.BackscatterBiharmonic(smag_constant=0.08, back_constant=1.1))
INFO: Logger initialized
INFO: Step: 25000, Time: 9.00e+07, KE: 5.35e-04, CFL: 0.048
CPU times: user 35.9 s, sys: 57.8 s, total: 1min 33s
Wall time: 27.5 s
Note how these are slightly slower than the baseline low-resolution model, but much faster than the high-resolution model.
See the parameterizations API section and code for examples of how these parameterizations are defined!
Compute similarity metrics between parameterized and high-resolution simulations¶
To assist with evaluating the effects of parameterizations, we include helpers for computing similarity metrics between model diagnostics. Similarity metrics quantify the percentage closer a diagnostic is to high resolution than low resolution; values greater than 0 indicate improvement over low resolution (with 1 being the maximum), while values below 0 indicate worsening. We can compute these for all diagnostics for all four simulations:
[9]:
def label_for(sim):
return f"nx={sim.nx}, {sim.parameterization or 'unparameterized'}"
sims = [high_res, backscatter, low_res, smagorinsky]
pd.DataFrame.from_dict([
dict(Simulation=label_for(sim),
**pyqg.diagnostic_tools.diagnostic_similarities(sim, high_res, low_res))
for sim in sims])
[9]:
| Simulation | Ensspec1 | Ensspec2 | KEspec1 | KEspec2 | ... | ENSfrictionspec | APEgenspec | APEflux | KEflux | APEgen | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | nx=256, unparameterized | 1.000000 | 1.000000 | 1.000000 | 1.000000 | ... | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| 1 | nx=64, BackscatterBiharmonic(Cs=0.08, Cb=1.1) | 0.617871 | 0.595677 | 0.719966 | 0.776835 | ... | 0.444769 | 0.222930 | 0.571082 | 0.448418 | 0.015557 |
| 2 | nx=64, unparameterized | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 3 | nx=64, Smagorinsky(Cs=0.08) | -0.049632 | 0.476905 | -0.242329 | -0.269631 | ... | -0.161524 | 0.069705 | -0.171567 | -0.296460 | 0.206204 |
4 rows × 19 columns
Note that the high-resolution and low-resolution models themselves have similarity scores of 1 and 0 by definition. In this case, the backscatter parameterization is consistently closer to high-resolution than low-resolution, while the Smagorinsky is consistently further.
Let’s plot some of the actual curves underlying these metrics to get a better sense:
[19]:
def plot_kwargs_for(sim):
kw = dict(label=label_for(sim).replace('Biharmonic',''))
kw['ls'] = (':' if sim.uv_parameterization else ('--' if sim.q_parameterization else '-'))
kw['lw'] = (4 if sim.nx==256 else 3)
return kw
plt.figure(figsize=(16,6))
plt.rcParams.update({'font.size': 16})
plt.subplot(121, title="KE spectrum")
for sim in sims:
plt.loglog(
*pyqg.diagnostic_tools.calc_ispec(sim, sim.get_diagnostic('KEspec').sum(0)),
**plot_kwargs_for(sim))
plt.ylabel("[$m^2 s^{-2}$]")
plt.xlabel("[$m^{-1}$]")
plt.ylim(1e-2,2e2)
plt.xlim(1e-5, 2e-4)
plt.legend(loc='lower left')
plt.subplot(122, title="Enstrophy spectrum")
for sim in sims:
plt.loglog(
*pyqg.diagnostic_tools.calc_ispec(sim, sim.get_diagnostic('Ensspec').sum(0)),
**plot_kwargs_for(sim))
plt.ylabel("[$s^{-2}$]")
plt.xlabel("[$m^{-1}$]")
plt.ylim(1e-8,2e-6)
plt.xlim(1e-5, 2e-4)
plt.tight_layout()
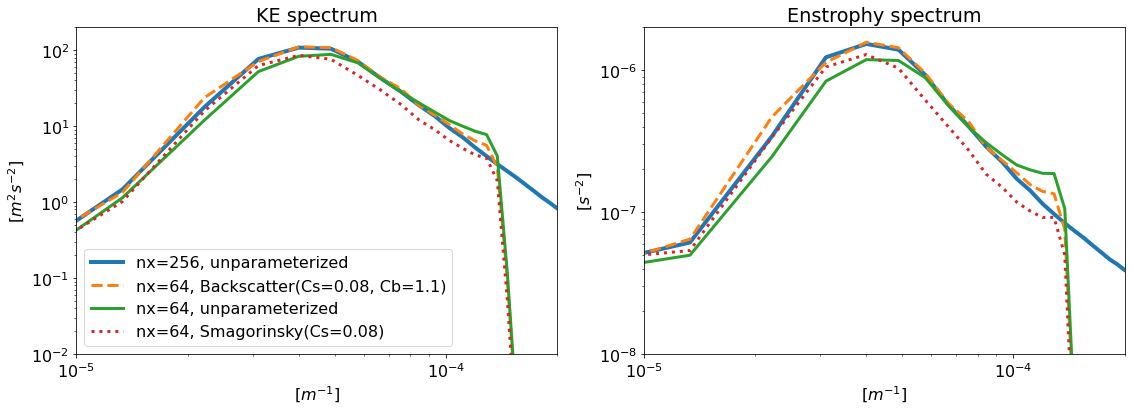
The backscatter model, though low-resolution, has energy and enstrophy spectra that more closely resemble those of the high-resolution model.